JE Mietus, C-K Peng, PCh Ivanov, AL Goldberger
Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, USA
|
This article originally appeared in Computers in Cardiology 2000,
vol. 27, pp. 753-756 (Piscataway, NJ: The Institute of Electrical and Electronics Engineers, Inc.). Please
cite this publication when referencing this material.
The software described in this article is freely available here.
|
Abstract:
We present a new automated method to diagnose and quantify obstructive sleep
apnea from single-lead electrocardiograms based on the detection of the
periodic oscillations in cardiac interbeat intervals that are often associated
with prolonged cycles of sleep apnea. This technique employs the Hilbert
transformation of the sinus interbeat interval time series to derive the
instantaneous amplitudes and frequencies of the series and calculates their
averages and standard deviations over a moving 5-minute window. We then apply a
thresholding technique and detect continuous sequences of those windows that
lie within threshold limits. When applied to the Computers in Cardiology sleep apnea test
data, our algorithm correctly classified 28 out of 30 cases (93.3%) of both
sleep apnea and normal subjects, and correctly identified the presence or
absence of sleep apnea in 14591 out of a total of 17268 minutes (84.5%) of the
data from the test set.
Introduction
Obstructive sleep apnea (OSA), the periodic cessation of breathing during
sleep due to intermittent airway obstruction, is a frequently undiagnosed
condition affecting millions of individuals worldwide, and is associated
with increased morbidity and mortality. Current technology for the diagnosis
of sleep apnea requires overnight monitoring of the patient in a specially
equipped sleep laboratory. Because of the expense and inconvenience of
standard polysomnographic recording, less costly and more easily implemented
techniques for detection of high-risk subjects would be desirable.
Our approach is based on the finding that OSA frequently alters healthy heart
rate dynamics. In normal healthy respiration, heart rate dynamics exhibit a
broadband, inverse power law spectral distribution [1]. In contrast,
during periods of prolonged OSA, the heart rate typically shows cyclic
increases and decreases associated with the apneic phase and the resumption
of breathing [2]. These cycles which tend to oscillate at a frequency
of between 0.01 and 0.04 Hz, are a distinctive feature of OSA not found during
normal respiration. We hypothesized that we could detect and quantify these
periods of high-density OSA by the fully automated identification of these
oscillatory dynamics in the RR interbeat interval series.
Methods
The heart rate oscillations accompanying prolonged OSA are transient, and
highly nonlinear and nonstationary, typically with varying amplitudes
and frequencies [Fig 1a]. These properties cause standard spectral
analysis techniques such as the Fourier transformation and autoregressive
methods to be ineffective in reliably detecting and localizing episodes of OSA.
The Hilbert transform is an analytical technique for transforming a time
series into corresponding values of instantaneous amplitudes and frequencies
[3, 4]. By using this technique to transform the RR interval
time series into its instantaneous amplitude and frequency components and
quantifying the mean values of these components and their stability over
time, it is possible to differentiate the periods of cyclic heart rate
behavior accompanying prolonged apnea from periods of normal respiration.
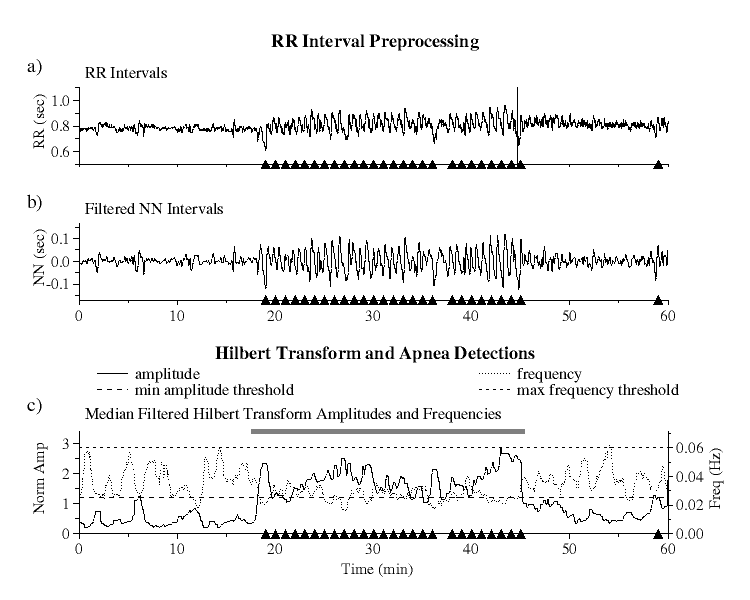 |
| Figure 1.
RR interval preprocessing and Hilbert transform amplitudes and
frequencies: a) raw RR interval series, b) bandpass filtered NN interval
series, and c) Hilbert transformed data showing minimum amplitude (long
dashed line) and maximum frequency (short dashed line) threshold limits.
Black triangles indicate actual apnea episodes. The grey bar indicates
the apnea detection based on our algorithm using the six parameters
described in the text.
|
RR Interval Preprocessing and Hilbert Transformation
Since the Hilbert transform requires a bandwidth limited signal, the RR
interval series must first be preprocessed to isolate frequencies of
interest.
To eliminate the effects of ectopic beats from the RR interval series
[Fig 1a], we select only normal sinus to normal sinus (NN) intervals as detected
by an automated beat detection and classification algorithm. Because QRS
detection algorithms may misclassify some ectopics as normal or may miss
some normal beat detections, we then apply a moving window average filter
to remove the remaining impulse noise due to these outliers as follows. For
each set of 41 contiguous NN intervals, a local mean is computed excluding
the central interval and those intervals which lie outside the range of 0.4
to 2.0 sec. The central interval is considered to be an outlier and is rejected
if its value lies outside of 20% of this mean. This test is applied to each
interval in the series and all such outliers are removed.
The filtered NN interval series is next linearly resampled at 1 Hz and
bandpass filtered by applying low and high pass filters [Fig 1b]. The
frequency range of the bandpass filter is selected to ensure that OSA
oscillations will not be filtered out. The low pass filter is a moving
window boxcar filter of width 5, giving a 3db cutoff at 0.09 Hz where the
value at each point in the series is replaced by the average value over
the window centered that at point. The high pass filter uses a local
detrending technique over a moving window of 81 data points, giving a
highpass 3db cutoff at 0.01 Hz. At each point in the series, the slope of
the regression line over a window centered at that point is computed, and
the value of this fit at the central point is subtracted from the actual
value at this point.
The Hilbert transform is then applied to the filtered NN interval series
and the instantaneous amplitude and frequency calculated at each point.
Since the amplitudes and frequencies of the transformed data exhibit
large fluctuations around their mean values, to eliminate this noise they
are each median filtered using a moving window of 60 points. To compensate for
the differing amplitudes of heart rate variability among different subjects,
the amplitudes are then normalized so that their average value over the
entire time series is equal to one [Fig 1c].
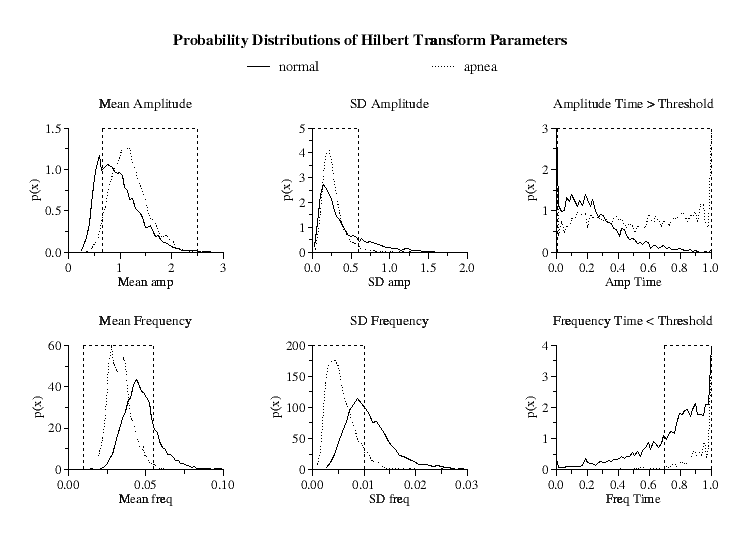 |
Figure 2.
Hilbert transform amplitude and frequency probability distributions
for the means, standard deviations and time within threshold limits for the
5-minute windows of all periods of normal respiration and high-density OSA.
Parameter limits indicated by dashed vertical lines optimize minute-by-minute
true positive and true negative apnea detections. Ordinates 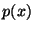 denote
the probability density, where denote
the probability density, where
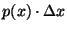 gives the
probability of a value occurring within a small region gives the
probability of a value occurring within a small region 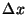 centered
at centered
at 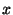 . The total area under each of the curves is equal to unity. . The total area under each of the curves is equal to unity.
|
Apnea Detection
The median filtered Hilbert amplitudes and frequencies are then analyzed
over 5-minute windows incremented for each minute. Six parameters are
calculated for each window increment: the average and standard deviation
of both the amplitudes and frequencies over the window and the fraction of
time that the amplitudes and frequencies are each separately within their
predefined threshold limits [Fig 1c].
To compare the distributions of these parameters for normal respiration and for
periods of prolonged OSA, we combined the PhysioNet Polysomnographic and Normal Sinus Rhythm (NSR) databases [5]
together with the Computers in
Cardiology (CinC) training dataset [5]. For the periods of normal
respiration, in addition to the eight hours of data from each of the 10 normal
subjects in the CinC training set, we selected from the 18 subjects in the NSR
database the six hours of lowest heart rate, corresponding to periods of sleep,
giving a total of 188 hours of normal respiration. For periods of prolonged
OSA, we selected from the the CinC training set and Polysomnographic database
those periods of time that were 15 minutes or greater in duration during which
there was at least 1 episode of apnea per minute, giving a total of 117 hours
of prolonged high-density OSA.
Since the probability distributions of these six parameters are different for
the periods of prolonged sleep apnea compared to periods of normal respiration
[Fig 2], by an appropriate selection of ranges for these parameters, it is
possible to selectively isolate windows of periodic NN interval behavior.
However, due to the overlap between the distributions, to discriminate
the periods of prolonged periodic heart rate dynamics in high-density OSA
from periods of intermittent heart rate periodicity found in normals, we
further require that each of these six parameters be continuously within
their specified limits (defined below) for a minimum of 15 minutes to qualify
as a detection of an apnea episode.
To determine the particular values of the Hilbert amplitude threshold and
the distribution parameter limits that give the best apnea detection, each of
these parameters were incrementally varied, and statistics were calculated
from multiple computer runs using data from the combined Polysomnographic,
NSR and CinC training set databases. The parameters selected were those that
gave the highest percentage of minute-by-minute true positive and true negative
apnea detections in these combined sets.
By this method, the ``optimal'' minimum Hilbert amplitude threshold was
determined to be a linear function of the minimum and maximum Hilbert
amplitudes given by
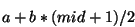
where 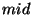 = the midpoint of the minimum and maximum amplitudes,
= the midpoint of the minimum and maximum amplitudes,
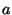 = -0.555 and
= -0.555 and 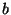 = 1.3.* The maximum Hilbert frequency
threshold was fixed at 0.06 Hz. The ranges for the six distribution
parameters giving the best detection ratios of all computer runs were
determined to be:
= 1.3.* The maximum Hilbert frequency
threshold was fixed at 0.06 Hz. The ranges for the six distribution
parameters giving the best detection ratios of all computer runs were
determined to be:
| mean amplitude (normalized) |
: |
0.65 |
- |
2.5 |
| standard deviation of amplitude |
: |
0 |
- |
0.6 |
| mean frequency (Hz) |
: |
0.01 |
- |
0.055 |
| standard deviation of frequency |
: |
0 |
- |
0.01 |
| time above amplitude threshold |
: |
0.006 |
- |
1.0 |
| time within frequency thresholds |
: |
0.7 |
- |
1.0 |
Finally, to separate subjects with severe OSA from normal subjects, we require
that a minimum of 5% of the total time of the RR interval series be detected
as apnea.
Results
Applying this technique to the CinC training set, we are able to correctly
classify 26 out of 30 (86.6%) of combined sleep and normal subjects, and
correctly identify the presence or absence of sleep apnea in 13985 out of
a total of 17045 minutes (82.1%) of the data.
When this technique is applied to the CinC test set, this algorithm correctly
classified 28 out of 30 (93.3%) of the subjects, and correctly identified the
presence or absence of sleep apnea in 14591 out of a total of 17268 minutes
(84.5%) of data from the test set.
Discussion
By using the Hilbert transformation of the RR interval time series, we have
shown that it is possible to detect prolonged episodes of high-density OSA
from single-lead electrocardiograms with a high degree of accuracy,
discriminating both subjects with OSA and determining the onset and offset
of prolonged high-density OSA. In addition, this technique is able to quantify
both the amplitude of OSA heart rate oscillations and their frequencies.
Improvements in filtering techniques and parameter adjustments may further
increase the efficacy of this technique.
Although this technique is ineffective in detecting the small number of sleep
apnea subjects who do not exhibit the oscillatory heart rate dynamics
commonly found in prolonged OSA, it may prove to be a cost-effective and
convenient means to screen for OSA and to monitor response to therapy. It
may also be useful in detecting and quantifying other pathologic heart rate
oscillations, such as those found in Cheyne-Stokes syndrome or periodic
breathing at high altitude [6], as well as fetal distress syndromes
[7].
Acknowledgements
This work was supported in part by grants from NIMH(MH54031),
NCRR(RR13622), the G. Harold and Leila Y. Mathers Charitable Foundation and
The Fetzer Institute. We thank G. B. Moody for helpful discussions.
References
- Kobayashi, M. and Musha, T., 1/f fluctuation of heartbeat period.,
IEEE Trans Biomed Eng, 29:456-457. 1982.
- Guilleminault, C. and Connolly, S. and Winkle, R. and Melvin, K. and Tilkian, A.,
Cyclical variation of the heart rate in sleep apnoea syndrome. Mechanisms, and usefulness
of 24 h electrocardiography as a screening technique, Lancet, 1:126-131. 1984.
- Ivanov, P. C. h. and Rosenblum, M. G. and Peng, C-K. and Mietus, J. and Havlin, S.
and Stanley, H. E. and Goldberger, A. L., Scaling behaviour of heartbeat intervals obtained
by wavelet-based time-series analysis, Nature, 383:323-327. 1996.
- Peng, C-K. and Mietus, J. E. and Liu, Y. and Khalsa, G. and Douglas, P. S. and Benson,
H. and Goldberger, A. L., Exaggerated heart rate oscillations during two meditation techniques,
Int J Cardiol, 70:101-107. 1999.
- PhysioNet: An NIH/NCRR Research Resource for Complex Physiologic Signals.
http://www.physionet.org
- Lipsitz, L. A. and Hashimoto, F. and Lubowsky, L. P. and Mietus, J. and Moody, G.
and Appenzeller, O. and Goldberger, A. L., Heart rate and respiratory rhythm dynamics on
ascent to high altitude, Br Heart J, 74:390-396. 1995.
- Kang, A. H. and Boehm, F. H., The clinical significance of intermittent fetal sinusoidal
heart rate, Am J Obstet Gynecol, 180:151-152. 1999.
Footnotes
- *
- Note: in the original published
version of this paper the values for
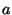 and
and 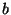 were incorrectly
given as
were incorrectly
given as 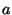 = 0.3 and
= 0.3 and 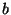 = 1.85.
= 1.85.

